RetroRules is a database of reaction rulesfor metabolic pathway discovery and engineering
In a nutshell
- RetroRules delivers a complete set of reaction rules spanning more than 15 000 biochemical transformations expressed at multiple levels of enzyme specificity.
- Reaction rules are generic descriptions of reactions expanding natural chemical diversity by predicting de novo reactions of promiscuous enzymes.
- Rules substrate specificity are encoded by considering the atomic environment around the reaction center at a given diameter.
- Rules are encoded in the community-standard SMARTS formalism.
- Rules metadata interrelates initial reactions, metabolites, EC numbers, and enzyme sequences.
- Rules are scored based on enzyme sequence availability, allowing prospective assessment and ranking of pathways.
- Current release of RetroRules is based on MNXref v3.0.
How to cite RetroRules?
Duigou T, du Lac M, Carbonell P, Faulon JL. RetroRules: a database of reaction rules for engineering biology. Nucleic Acids Research, 2019. | doi: 10.1093/nar/gky940 | PMID: 30321422Reaction rules generation
Reaction rules were generated using the procedure outlined below:
- Extract reaction information from metabolic databases. Filter out reactions that miss any structure from amongst involved compounds.
- Remove reactions that do not trigger the modification of substrate (e.g. passive transport) or that involve compounds not fully characterized (e.g. R-groups).
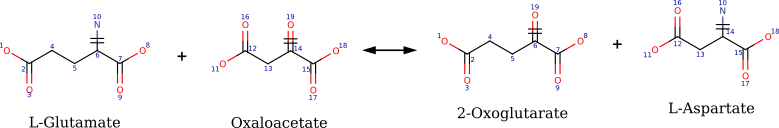
- Identify the reaction center (i.e. subpart(s) of substrate(s) that are transformed) based on an atom-atom mapping between substrates atoms and product atoms (AAM). Figure below shows reaction 2.6.1.1 with atom mapping, reacting atoms are those labelled 6, 10, 14 and 19.
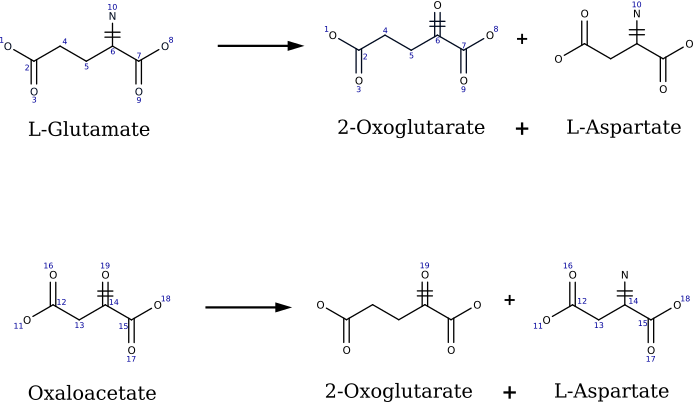
- Decompose multi-substrate reactions into mono-substrate component. There are as many components as there are substrates and each component gives the transformation between one substrate and the products. Each product must contain at least one atom from the substrate according to the AAM. This strategy enforces that only one substrate can differ at a time from the substrates of the reference reaction when applying the rule. Reaction decomposition to mono-substrate component are considered for both direction of reactions, enabling utilization of reversed rules for retrosynthesis application. Figure shows the mono-substrate component generated from reaction 2.6.1.1.
- Optionally, substrate compounds that are cofactors (such as water, CO2, ATP, NADP, ions, ...) can be ignored until the end of the procedure under the assumptions that such metabolites are available in the cell and that there is no gain to consider promiscuity on them. RetroRules' current release does perform a cofactor removal (list provided as SI of database paper).
- Compute the reaction rules using the reaction SMARTS formalism for each mono-substrate component. Perform rule SMARTS generation considering different diameters around the reaction center by removing from the components atoms that were not in the spheres around the reacting atoms. Figure shows reaction rules generated when considering three differents diameters around the reacting atoms of L-glutamate ("Substrate"). RetroRules' current release provides reaction rules for diameter 2 to 16.
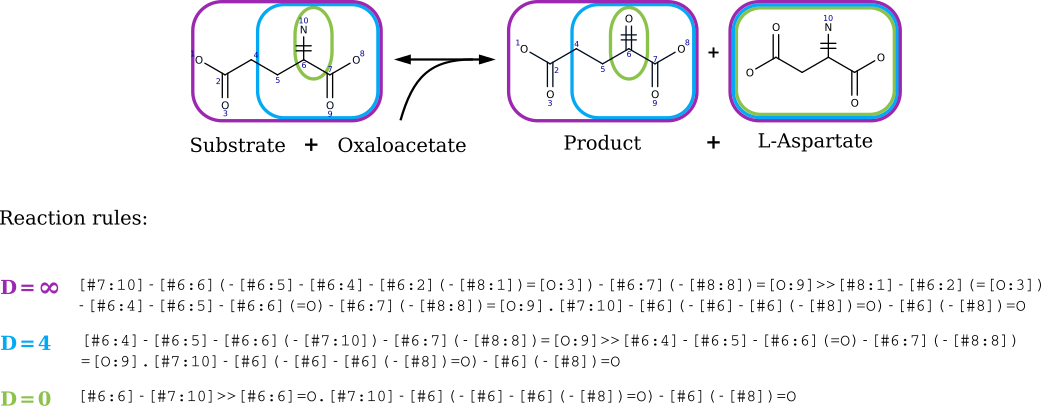
An in depth description and validation of the generation process are described in Delépine et al., 2018.
Reaction rule scoring system
Reaction rules are scored based on enzyme sequence availability, allowing prospective assessment and ranking of pathways.
Scores associated to reaction rules should be regarded a penalty score: a penalty score of 0 is the best possible value, while a higher penalty (>0) implies a decrease in the certainty of sequence availability.
More details on the score computation and investigation are described in Delépine et al., 2018.
RetroRules database schema
Data used in RetroRules are available as a SQLite file. Figure below depict the SQL schema in used by RetroRules.
At the center of the schema is the rules table that contains the information to uniquely describe a reaction rule from a mono-component reaction, where one rule is associated with a given substrate from a given reaction at a given diameter. However each rule can have multiple products, described in the rule_products table. The SMARTS and SMILES description of these rules can be found in the smarts and smiles tables respectively since a given SMARTS or SMILES description of a rule can apply to multiple reactions, substrates and diameters. The rest of the tables are meta-informations extracted from source databases. For more information feel free to contact us.
Online rule generator
The online rule generator allows one to generated custom rules using a reaction SMILES depiction or a RXN MDL file describing the reaction template to be used.
The same procedure than described in the "Reaction rules generation" section is used to generate the rules, with 2 exceptions. First, all structures of the inputed reaction are considered as primary compounds, i.e. no filtering will be attempted to remove cofactors. We believe that the role importance of each structure involved in a reaction is up to the user. To help in this, the second difference is that the custom rule generator allows the use of unbalanced reactions, i.e. reaction where the number of atoms different between left and right hand sides.
REST API
See here for technical documentation and examples on the REST API.