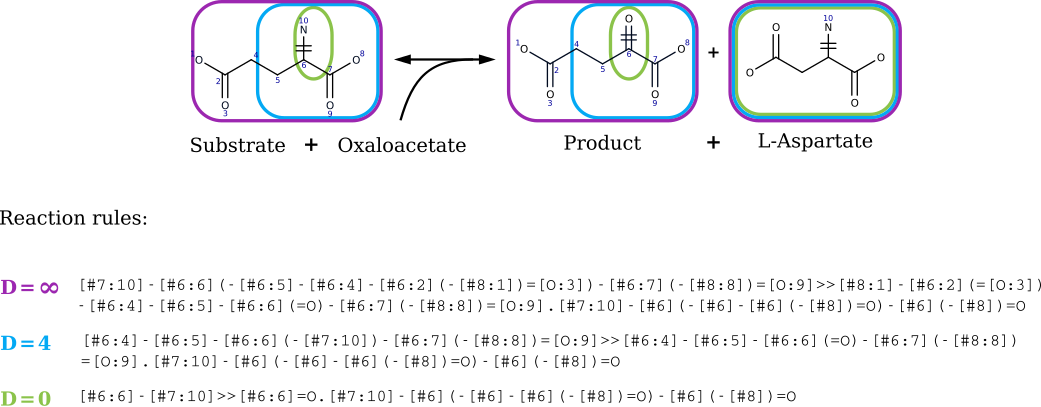At a glance
- Templates in community‑standard SMARTS syntax.
- Specificity via a tunable radius around reacting atoms.
- Linked metadata: source reactions, compounds, EC numbers, sequences, structures.
- Sequence‑aware scoring to prioritize plausible routes.
Datasets
- From MetaNetX, Rhea and USPTO-50k.
- ~400k templates from >40k biochemical reactions.
- ~1M templates from >90k reactions.
- ~140k templates associated with sequences.
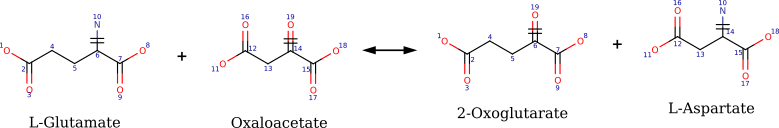
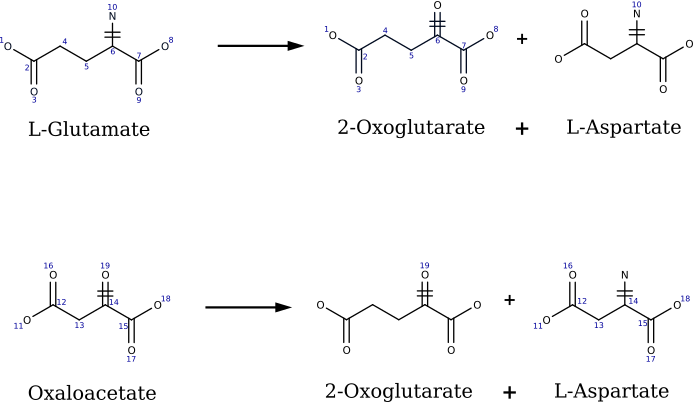
What is a reaction template?
- Concept
- A reaction template abstracts a biochemical transformation by focusing on the atoms that change during the reaction (reaction center) and a surrounding neighborhood (radius).
- Radius
- By varying the radius, the same underlying chemistry can be represented at multiple levels of enzyme specificity.
- SMARTS
- Templates are stored as reaction SMARTS, which compactly encode the left and right atomic patterns that define the reaction.
Using Templates in RetroRules
- Search by SMARTS, EC, template ID, or radius; filter by dataset.
- Inspect details: source reactions, EC numbers, compounds, sequences.
- Follow link‑outs to external registries (Rhea, MetaNetX, ChEBI, UniProt).
- Export filtered sets for downstream workflows.
Reaction rules generation
Reaction rules were generated using the procedure below. Click a step on the right to see details.
1. Extract reaction information from metabolic databases. Filter out reactions that miss any structure among involved compounds.
Sequence-aware scoring
Biochemical templates are associated with an enzyme evidence score gathered from source reactions, UniProt sequences and EC annotations. Higher is better; near 0 indicates less confidence in enzyme availability or assignment. This helps rank alternative routes during pathway search. Non-biochemical templates (namely USPTO) do not have this scoring.
See Duigou et al., 2019 for details.
Working across multiple datasets
RetroRules aggregates templates and reactions from multiple sources (e.g., MetaNetX, Rhea, USPTO). A given template may appear in multiple template sets, indicating it is supported by reactions from different datasets. Its provenance is preserved and shown in the UI. MetaNetX Rhea USPTO
API quickstart
Search templates:
GET /api/templates?ec=1.2.1&radius=4
Retrieve a template summary:
GET /api/templates/<TEMPLATE_ID>/summary
See the API Docs for full parameter details and examples.
Citing RetroRules
If you use RetroRules in your research, please cite:
Duigou T, du Lac M, Carbonell P, Faulon J-L, RetroRules: a database of reaction rules for engineering biology. Nucleic Acids Research, 2019. doi: 10.1093/nar/gky940
FAQ & glossary
Template ID vs. Reaction ID?
A Template ID identifies a unique SMARTS pattern. A Template aggregates multiple source reactions; Reaction IDs refer to individual reactions in the datasets.
Why multiple EC numbers?
A template may be supported by reactions annotated with different ECs (or partial ECs), reflecting enzyme diversity.
Can templates be reversed?
No—templates are asymmetric, the left side represents the substrate pattern to be matched, while the right side formalizes the reorganization of atoms to be applied to form products. However, templates for both forward and reverse directions are generated for all reactions.
Key terms
- AAM: atom–atom mapping between substrates and products.
- Radius (or Diameter): radius (in bonds) of neighborhood kept around reacting atoms.
- Promiscuity: enzyme activity on non-native substrates predicted by templates.